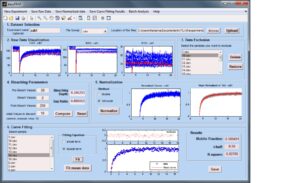
EasyFRAP assists quantitative and qualitative analysis of FRAP data.
The user can handle simultaneously large datasets of raw data, visualize fluorescence recovery curves, exclude low quality data, perform data normalization, extract quantitative parameters, perform batch analysis and save the resulting data and figures for further analysis.
EasyFRAP is implemented as a single-screen Graphical User Interface and is highly interactive, as it permits parameterization and visual data quality assessment at various points in the analysis. EasyFRAP is free software, available under the General Public License version 3 (GPL v3).
Software:
To use easyFRAP, you have 2 options:
- Standalone version: Please first download and install MATLAB Runtime from this link (check the version in the Comments of the Table). This is required for standalone applications to run outside MATLAB. Then download the appropriate version of easyFRAP and run easyFRAP.exe (Windows) or easyFRAP.app (Mac). Here you can also find a copy of the GPL license v3.
| Download easyFRAP | Compatibility | Comments |
| easyFRAP_Win.zip | Windows, 64 bit | Download R2016a (9.0.1) for Windows |
| easyFRAP_Mac.zip | Mac OSX 10.11 or later | Download R2015b (9.0) for Mac |
- Source files: download the MATLAB source code for Windows or Mac and run them through MATLAB. This requires a version of MATLAB installed on your computer.
Updates:
- Updated version uploaded on 19/6/2017. Please download the new version using the above links.
- Contact us at lygerou{at}upatras{dot}gr for feedback
- If you are experiencing an upload error with .txt or .csv files using Matlab Runtime 9.0 or later please read here.
Test data:
To get started with easyFRAP, experiment with these test datasets:
- single experiment – Download (.csv files)
- multiple experiments (for the Batch analysis feature of easyFRAP) – Download
Documentation:
- easyFRAP Quick Start Guide (step by step guide with screenshots) – Download
- detailed easyFRAP manual – Download
The paper describing easyFRAP can be accessed through Oxford journals here.
If you use our tool please cite:
Rapsomaniki MA, Kotsantis P, Symeonidou IE, Giakoumakis NN, Taraviras S and Lygerou Z (2012). easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of Fluorescence Recovery After Photobleaching data. Bioinformatics. 28(13):1800-1801
NEW: EasyFRAP-web
EasyFRAP became web-based! Visit EasyFRAP web and see the publication here.
Estimation of protein diffusion and binding parameters:
For inference of kinetic parameters from FRAP data (diffusion coefficient, bound fraction and residence time) see
Rapsomaniki, M. A., Cinquemani, E., Giakoumakis, N. N., Kotsantis, P., Lygeros, J., & Lygerou Z. (2015). Inference of protein kinetics by stochastic modeling and simulation of fluorescence recovery after photobleaching experiments. Bioinformatics, 2015 Feb 1;31(3):355-62.
contact us for more details at lygerou{at}upatras{dot}gr
